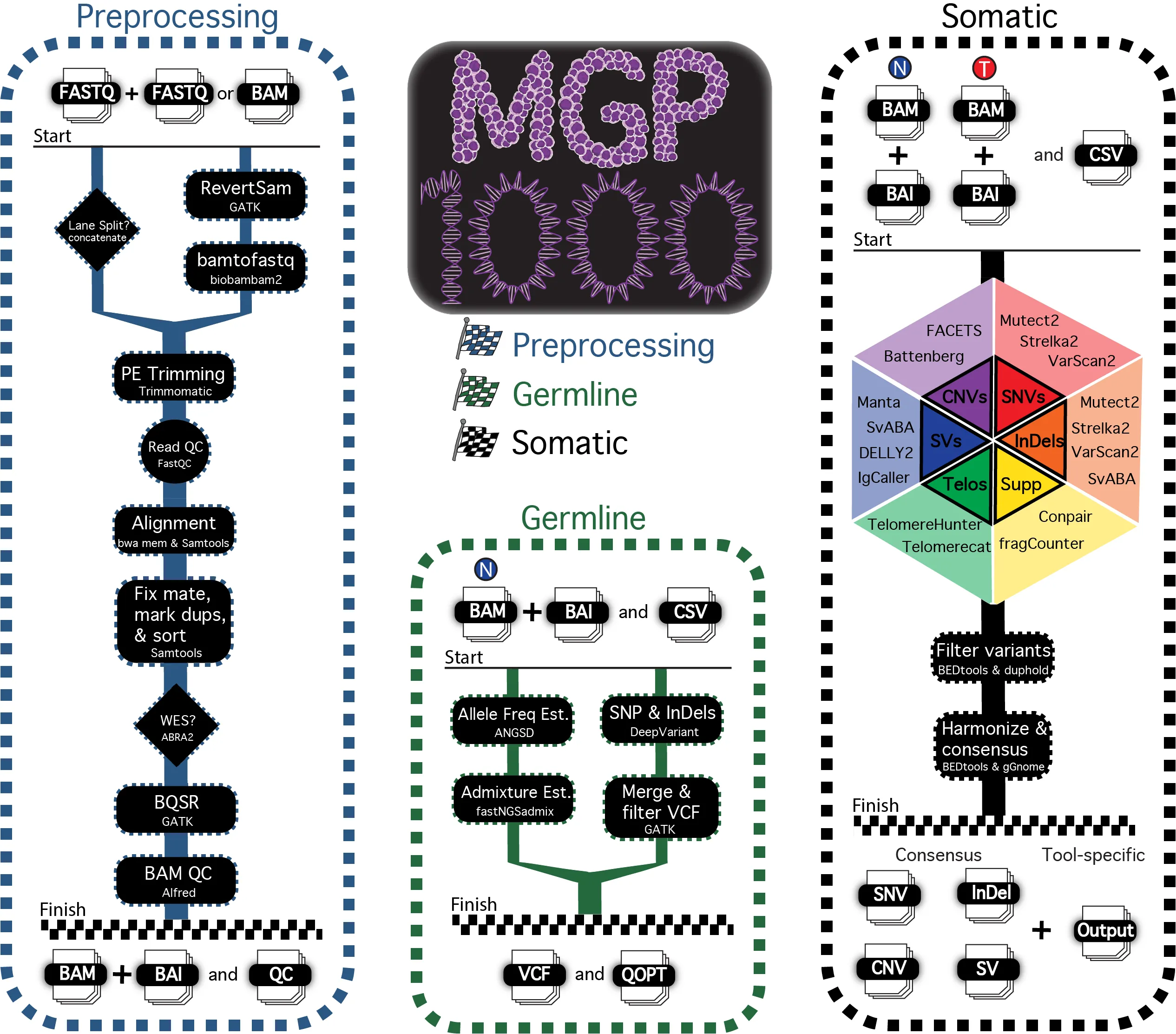
Introduction
In order to analyze over one thousand matched tumor/normal whole-genome samples across multiple data centers in a consistent manner, a pipeline was created that leverages the workflow management, portability, and reproducibility of Nextflow in conjuction with Singularity.
The MGP1000 offers a singular, consistent, automated workflow that is portable and reproducible across different data centers with little effort or issues stemming from environment incompatibilities, version inconsistencies, missing dependencies, or a need to setup the necessary tools on a user’s HPC infrastructure.
Pipeline Workflow
The entire pipeline is divided into 3 modules: Preprocessing, Germline, and Somatic